BackgroundOur laboratory studies beneficial associations between microbes and plants. We mainly focus on two symbioses: associations between plants and nitrogen-fixing bacteria (diazotrophs) and associations between plants and mycorrhizal fungi. We seek to understand the nature of microbial signals and the signaling pathways that control the perception of these signals in plants and fungi. We try to understand how these molecular mechanisms evolved in plants, bacteria, and fungi. We try to use this mechanistic and evolutionary knowledge to improve the benefits of microbes to crops, particularly the benefits of biological nitrogen fixation to non-leguminous plants for food, feed, fiber, and biofuel production.
|
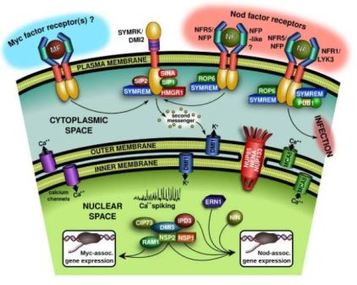
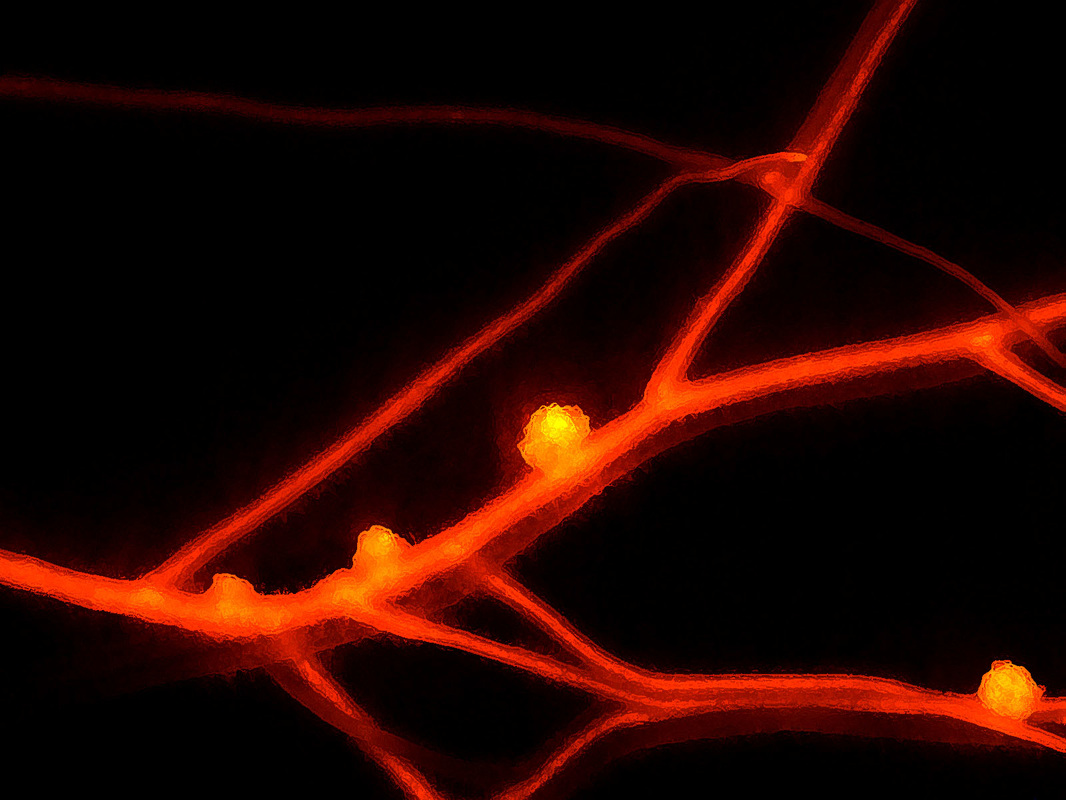
Plant-microbe and microbe-microbe “sweet talk” in symbiotic associations.Dr. Ané started his academic career under the mentoring of Dr. Frédéric Debellé and Dr. Jean Dénarié in Toulouse, France, trying first to determine the structure of lipo-chitooligosaccharides (LCOs), also called Nod factors and the role of nod genes in LCO synthesis in Mesorhizobium ciceri, the symbiont of chickpea. For his Ph.D. with Dr. Charles Rosenberg and Dr. Jean Dénarié, Dr. Ané transitioned to the plant side by mapping and identifying plant genes required for LCO signaling in the new model legume at the time, Medicago truncatula. Mutants unable to transduce the LCO signal failed to associate with rhizobia and arbuscular mycorrhizal (AM) fungi, revealing the existence of a Common Symbiotic Pathway (CSP) between the two symbioses. At this time, very few tools were available in M. truncatula, so Dr. Ané got involved in developing the first genetic map and other essential tools for this model legume. Dr. Ané continues this line of work during his postdoc at the University of California - Davis with Dr. Douglas Cook, which led, in particular, to the identification of the DMI1 and DMI3 (Does not Make Infection) genes, which were both published in Science. DMI1 had similarities with cation channels. DMI3 is a calcium/calmodulin-dependent kinase (CCaMK). |
Dissecting the legume Common Symbiosis Pathway (CSP)To further characterize the LCO signaling pathway in M. truncatula, we localized the DMI1 protein to the nuclear envelope. We provided evidence that DMI1 is a calcium-regulated calcium channel essential for nuclear oscillations of the calcium concentration called “calcium spiking”. We identified proteins interacting with DMI2 and DMI3. A key DMI2 interactor is a 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR1) that produces mevalonate, and we showed that HMGR activity is necessary and sufficient to activate calcium spiking. We first identified the transcription factor IPD3 (Interacting Protein of DMI3), later called CYCLOPS in Lotus japonicus, which turned out to be the primary output of the DMI3/CCaMK calcium spiking decoder.
|
Development of innovative tools for the model legume Medicago truncatula.
|
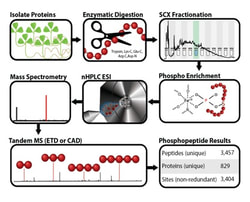
In collaboration with Dr. Michael Sussman, Dr. Joshua Coon, and Dr. Sushmita Roy at UW-Madison, we developed innovative techniques to characterize the metabolome, proteome, and phosphoproteome of M. truncatula. Such projects led to the identification, for instance, of EPP1 (Early Phosphorylated Protein 1), which is also an essential component of the CSP required for calcium spiking activation. We also worked with Dr. Lingjun Li (UW-Madison) on metabolite and peptide imaging by MALDI-TOF and Dr. Matias Kirst (University of Florida) for a single-nuclei RNA-seq analysis of nodule development.
|
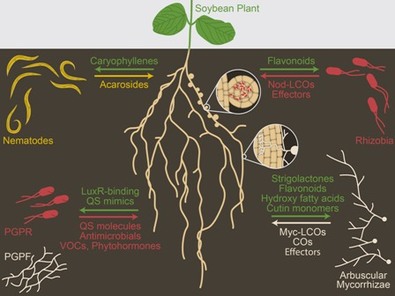
The evolution of plant-microbe symbiosesAlthough our initial work was focused on M. truncatula symbiosis with rhizobia and AM fungi, we became interested in understanding the evolution of these signals and signaling pathways.
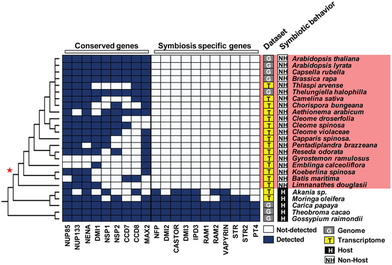
From legumes to liverworts and charophyte algae. AM associations occur in legumes and the vast majority of land plants. We analyzed the conservation and evolution of the CSP between M. truncatula and other legumes, other eudicots, monocots , mosses hornworts and liverworts to charophyte algae . In most of these studies, we combined phylogenomic approaches with genetic and functional assays. This work also paved the way for engineering the root nodule symbiosis in non-legumes such as poplar. The role of LCOs across and beyond symbioses. We suggested the presence of LCOs in exudates which was later demonstrated by Dr. Jean Dénarié. We then showed that ectomycorrhizal fungi also produce LCOs and that hosts like poplar (Populus sp.) elicit calcium spiking in response to these signals in a CSP-dependent manner. Finding LCOs in such diverse fungi, we explored the entire fungal kingdom. We discovered that most fungi produce LCOs and respond to them in a dose-dependent manner, suggesting that LCOs could be equivalent to bacterial quorum-sensing molecules in fungi. |
|
|
Increasing the benefits of microbes to agricultureTesting the efficiency of microbial inoculants.
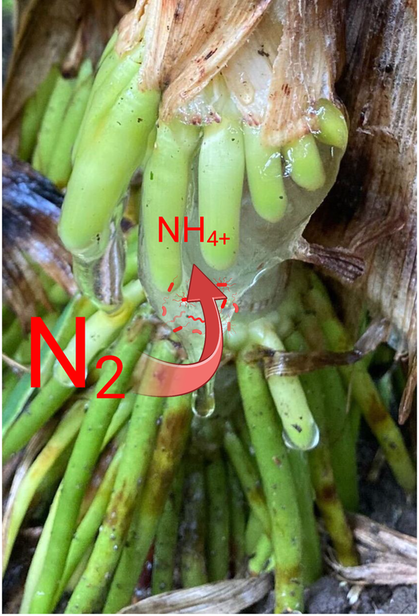
In collaboration with colleagues from the Agronomy and Plant Pathology departments, we worked on various applied projects on soybean rhizobia, Fusarium, Sclerotinia, nematodes, and herbicides. We generally brought my expertise in microbiology and molecular techniques to answer questions directly relevant to the field and also learned to design and conduct my field experiments. We are also working in collaboration with the Valent BioSciences company on better understanding the molecular basis (glomalin) for the benefits of AM fungi to host drought tolerance and soil health. Engineering of associative “ammonium excreters” for non-leguminous crops and, in particular, cereals. Plants host a wide range of diazotrophs in their rhizosphere, but these bacteria release minimal amounts of nitrogen to plants. Using genetic engineering, we developed “ammonium excreting” strains of Azotobacter vinelandii and Klebsiella strains that fix nitrogen even in the presence of nitrogen in their environment and release significant amounts of nitrogen to cereals crops. Some of this work is in collaboration with the Pivot Bio company. Genetic approaches to force diazotrophs to fix more nitrogen and release ammonium are extremely promising but lead to a decreased fitness of engineered bacteria. We are pursuing synthetic biology approaches in which ammonium excretion depends on signals produced by host plants. We are exploring the role of bacteria-bacteria and bacteria-fungi interactions using synthetic communities to identify microbial competitors and helpers of the engineered diazotrophs in the rhizosphere. Exploring and using natural diversity in cereals for better plant hosts. Towards the same goal of improving the benefits of biological nitrogen fixation for cereal crops, we characterized corn landraces from Southern Mexico that associate can acquire about half of their nitrogen from the air by hosting diazotrophic communities in a mucilage produced by aerial roots. We currently continue characterizing the genes and mechanisms allowing aerial root development and mucilage production in a project funded by USDA. Given that this trait was initially identified and used in corn landraces by Indigenous communities in Southern Mexico, we are also exploring avenues to ethically and collaboratively work with these communities. In collaboration with Dr. Vermerris (University of Florida), we also identified sorghum accessions with the same trait. We are working on mapping and introgressing this trait into accessions adapted to a broader range of environments for both corn and sorghum. |